Experiment Configuration¶
NXRefine uses a hierarchical directory structure for each experiment to store both the raw data generated on an x-ray beamline and processed data generated by the data reduction workflow. An ‘experiment’ in NXRefine comprises measurements on a set of samples that are logically grouped together, often because they are performed within a specific time period and/or share calibration files. Plugins to the NeXpy GUI are used to facilitate the creation of both the directory hierarchy and the associated data files.
Data that are processed by the NXRefine package are stored as HDF5 files that conform to the NeXus format, which is an international standard for the storage of x-ray and neutron scattering data. There is a single NeXus file associated with each measurement, which consists of one or more rotation scans usually at a single temperature or other parametric variable. This file contains all the information required for a complete analysis, including external links to the raw data, beamline monitors, powder diffaction calibrations, and metadata generated by the data reduction workflow.
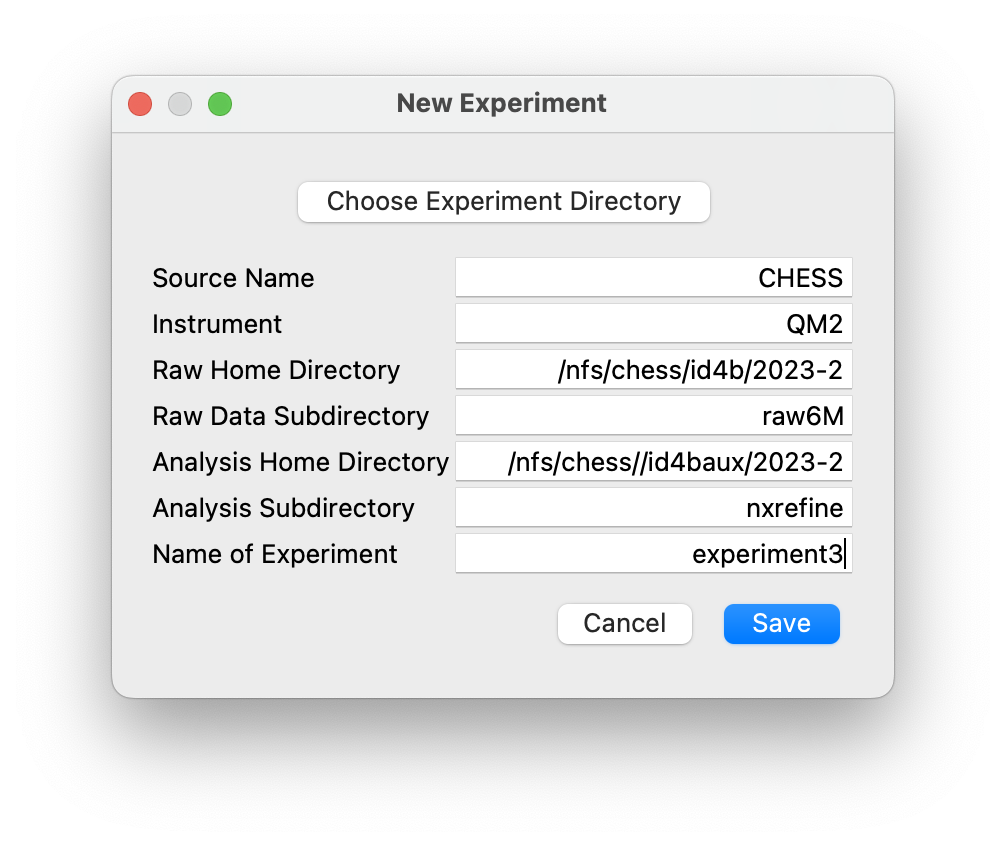
In this section, we will describe the NXRefine directory framework, explaining where the NeXus files are stored and how they are linked to the reduced data transformed into reciprocal space. At the beginning of each new experiment, the GUI dialogs launched from the Experiment Menu can be used to create the experiment directories, NeXus file templates, and new NeXus files based on those templates to contain scan data.
Experiment Layout¶
In NXRefine, it is assumed that all the files associated with a
particular experiment are stored in a single directory. At synchrotron
x-ray facilities, it is common to schedule all the measurements
associated with a particular proposal together, associated with a
proposal number and/or run cycle. For example, on beamline 6-ID-D at the
Advanced Photon Source, measurements resulting from Proposal No.
GUP-75969 may be scheduled in a specific run cycle, say 23-1, and stored
in, e.g., /data/6-id-d/GUP-75969-23-1. This directory should
contain sub-directories that conform to a particular layout, with
calibration files stored in the directory calibrations, NeXus files
containing measurement templates stored in configurations, settings
and log files stored in tasks, and experimental data from each
sample stored in separate directories with a name that is typically
derived from the chemical formula or a commonly used abbreviation. It is
common to test and/or measure multiple crystals with the same chemical
formula, so each of the sample directories contain sub-directories for
each measured crystal. These contain the NeXus files for each scan and
linked sub-directories containing the raw data.
Here is the structure of a possible experiment directory. Most of the
names in this example are chosen to be generic, i.e., they will be
different for every experiment. The only exceptions are the files in the
tasks directory, which are set automatically by NXRefine.:
experiment
└── tasks
├── nxdatabase.db
├── nxlogger.log
├── settings.ini
└── calibrations
└── powder_calibration.tiff
└── configurations
├── configuration.nxs
└── sample1
└── label1
├── sample1_100K.nxs
└── 100K
├── f1.h5
├── f2.h5
├── f3.h5
├── f1_transform.nxs
├── f2_transform.nxs
├── f3_transform.nxs
└── transform.nxs
├── sample1_200K.nxs
└── 200K
├── ...
└── sample1_300K.nxs
└── 300K
├── ...
└── label2
└── sample1_300K.nxs
└── 300K
├── ...
├── sample2
├── sample3
The goal of this directory structure is for all the data and metadata required to analyze the results to be stored in an easily accessible location, although not all files are required by NXRefine to be present. For example, the powder calibration files can be imported from any location.
If an instrument does not exclusively use NXRefine for its data
reduction, it is possible that an experiment directory already exists.
If so, in order to avoid any interference with other instrument files,
it is possible to create the above directory structure within a
sub-directory, called nxrefine, contained within the existing
experiment directory.:
experiment
└── nxrefine
├── tasks
├── calibrations
├── configurations
├── scripts
├── sample1
└── sample2
.
.
.
Note
The name of this sub-directory is defined in the server
settings, which are described below. It is strongly recommended that it be called by the default name, i.e.,
nxrefine to facilitate parsing of the directory tree.
Experiment Sub-Directories¶
- tasks
The
taskssub-directory contains a number of files used by NXRefine to store default settings, workflow logs, and a MySQL database for recording the status of each workflow component. The files in this directory are created automatically by NXRefine and should not be touched. NeXpy GUIs are used to inspect their contents.- calibrations
The
calibrationssub-directory is designed to contain either the TIFF or CBF files generated by measurements of a calibrant powder, or a file, usually with extension.poni, containing the instrument parameters calibrated using the PyFAI module. The workflow includes a GUI for performing PyFAI calibrations directly on powder calibration image files, with the results stored in the NeXus files (described below). The files don’t have to be stored in this sub-directory, but if they are in another location, it is recommended to copy them here for completeness. If the calibrations have been performed by another package, the parameters can be imported directly from a PONI file.- configurations
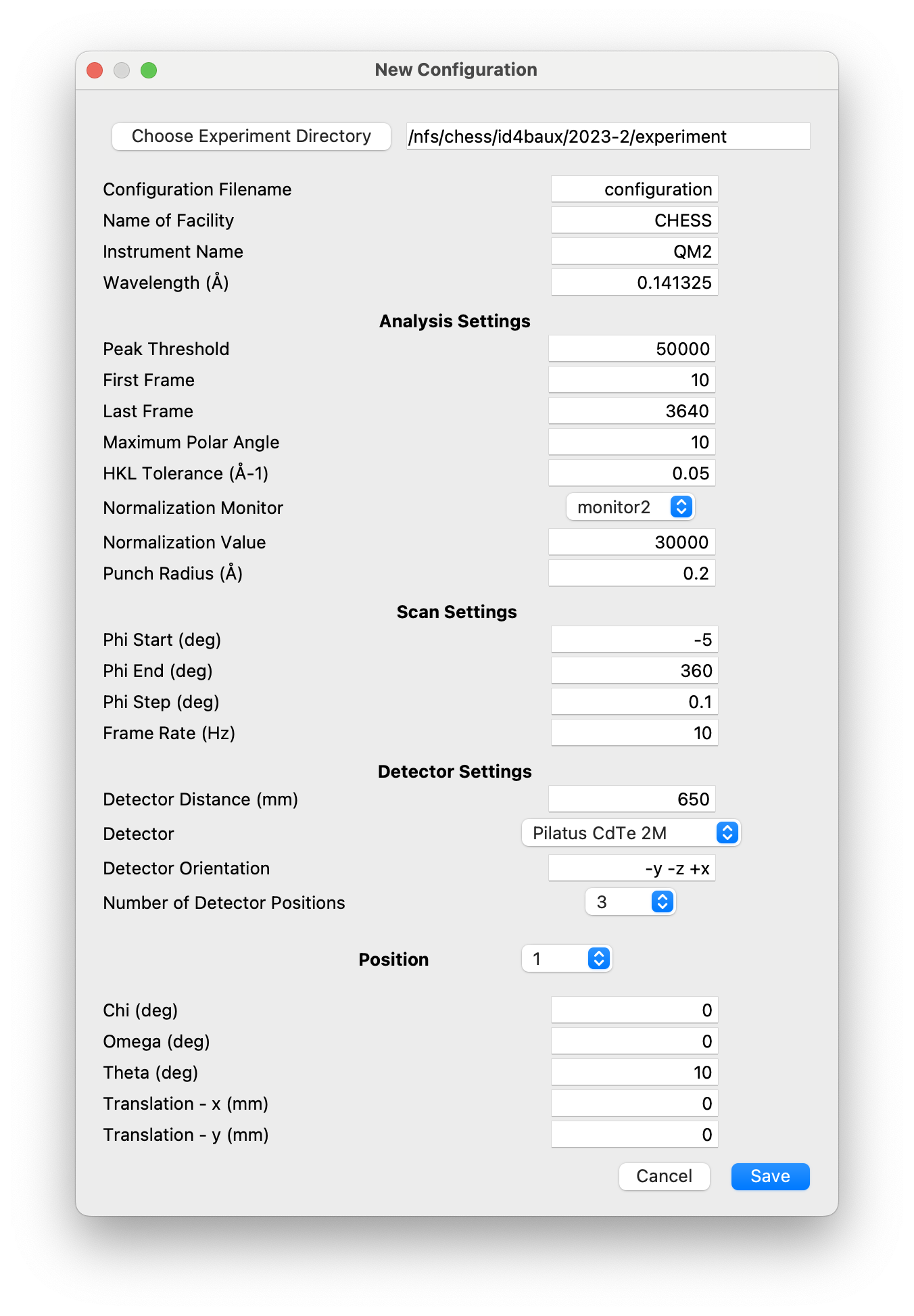
The
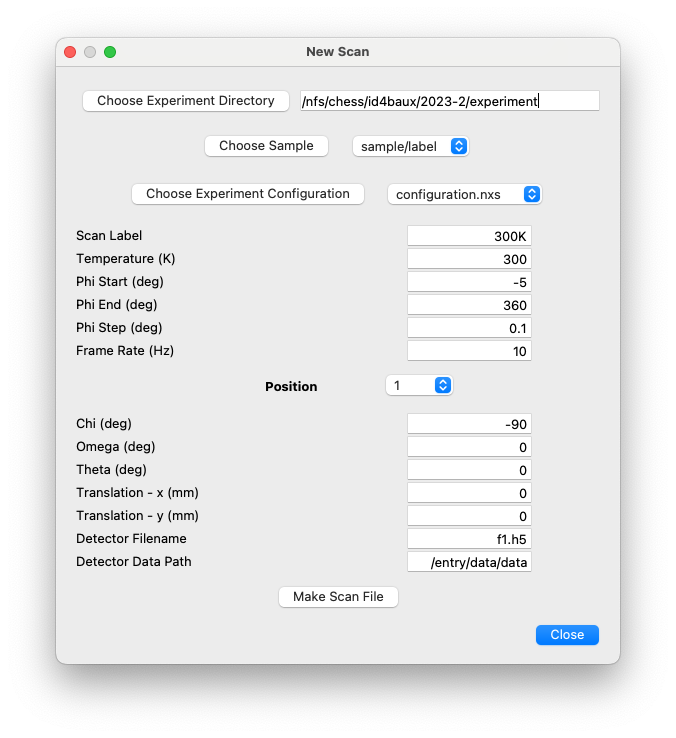
configurationssub-directory contains NeXus files that act as templates to be used when creating the files used to store the scan results. There should be a separate template file for each new experimental configuration, .e.g., with a different wavelength or detector distance. If multiple sample rotations are to be performed with different detector translations and/or goniometer angles, the corresponding template files will have entries for each scan containing pre-defined values of the scan variables. These files are initialized by a NeXpy GUI dialog.
Note
On QM2 at CHESS, it is usually only necessary to create template files with a single entry, since the number of rotation scans is not always pre-determined. When the scans are imported, additional entries are automatically added with the goniometer angles updated with the values in the corresponding scan SPEC file.
- scripts
The
scriptssub-directory is not used directly by NXRefine, but is created by theNew Experimentdialog described below. It is designed to store macros for use during an experiment.- sample
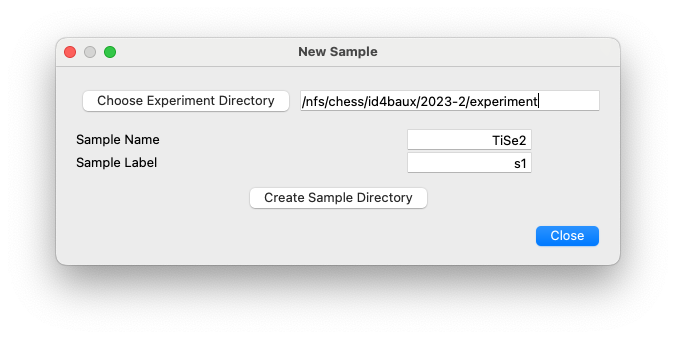
The
samplesub-directories are typically named after a common abbreviation or chemical formula of the measured sample (e.g.,TiSe2). Within each sample directory are one or more directories usually corresponding to different crystals, specified by unique labels typically provided by the crystal grower. It is common in these experiments to screen a number of crystals before selecting one for further measurements, in which case many of these directories would only contain a single scan.Within each
labeldirectory, there are one or more directories that are named after the parametric variable being modified between each set of rotation scans, e.g.,100K. Thesescandirectories contain the raw data in HDF5 files, typically with extension.h5. Each one of these.h5files contain the raw data from a single rotation scan stacked into a single HDF5 array. It is common to perform three sample rotations, which are then stored inf1.h5,f2.h5, andf3.h5, but any number is possible. Thescandirectories also contain other files produced during the data reduction procedure, such as data transformed into reciprocal space coordinates or pair-distribution functions.For each of these
scandirectories, there is a corresponding NeXus file that is named as, e.g.,sample_scan.nxs, wheresamplemust be the name of thesampledirectory andscanshould be the name of the directory containing the raw data. These NeXus files contain external links to the much larger files stored in thescandirectories. By opening them, the user has access to all the data and metadata associated with a particular scan, since external links, if they are available, will appear to be part of the file.
Note
External links are defined by the file name and internal path
to the required HDF5 field. If the file and/or field are not
available, the NeXus file can still be opened, but the
corresponding data cannot be viewed. The file name is stored
as a relative file path, so the NeXus file and a subset of
the files in the scan directory can be moved to another
location if, for example, access to the raw data is no
longer necessary.
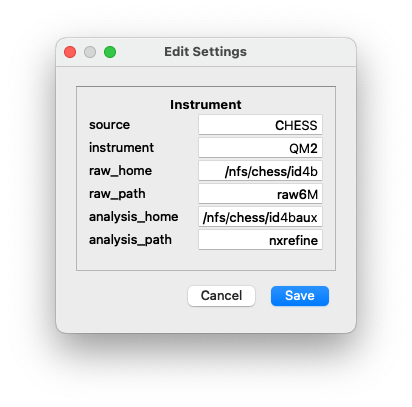
Instrument Setup¶
The experiment directory layout can be created automatically using GUI
dialogs in the NeXpy “Experiment” menu. Before using them, it is
important to have initialized the default instrument parameters using
the “Edit Settings” dialog of the NeXpy “Server” menu, or at the
command line using nxsettings -i.
The instrument settings provide information on the directories, in which
both the raw data and the NXRefine directory tree are located. It is
quite common for the raw data to be collected as a set of image files,
typically TIFF or CBF files. These are usually not stored in the
experiment directories described in the previous section, and may be in
read-only directories. To allow for the input of such files, NXRefine
defines two sets of paths; one to the ‘raw’ data and one to the
NXRefine (or ‘analysis’) directories. It is assumed that the
experiment names, e.g., GUP-75969-23-1, are the same in both
locations, although alternative methods of linking the ‘raw’ and
‘analysis’ paths could be defined in the customized beamline classes
described later.
For example, at CHESS, the ‘raw’ and ‘analysis’ paths are defined in parallel directory trees as follows (with generic experiment names):
/nfs/chess/id4b /nfs/chess/id4baux
├── 2023-1 ├── 2023-1
├── 2023-2 ├── 2023-2
└── 2023-2 └── 2023-2
├── experiment1 ├── experiment1
├── experiment2 ├── experiment2
└── experiment3 └── experiment3
└── raw6M └── nxrefine
├── sample1 ├── sample1
├── sample2 ├── sample2
└── sample3 └── sample3
└── label1 └── label1
├── 100 ├── 100
├── 200 ├── 200
└── 300 └── 300
Here is a list of instrument parameters.
- source
This is the name of the synchrotron source, at which the instrument is located. This will be stored in the NeXus files during the data reduction, but is not otherwise used.
- instrument
This is the name of the instrument. If a customized beamline package is to be imported, this must correspond to the instrument name used in the package.
- raw_home
This is the home directory, in which the experimental raw data are stored. In the above example, this could be
/nfs/chess/id4b/2023-3.- raw_path
This is the path within the experiment directory to the sample directories. In the above example, this would be
raw6M.- analysis_home
This is the home directory, in which the data are analyzed. In the above example, this could be
/nfs/chess/id4baux/2023-3.- analysis_path
This is the path within the experiment directory to the NXRefine sub-directories. In the above example, this would be
nxrefine.
On Sector 6 at the APS, the images are automatically stacked as HDF5
files and saved in the analysis directories as f1.h5, f2.h5,
etc, so the paths to the raw data are not required and can be left
blank. The ‘analysis_path’ field is also blank, since the sample
directories are at the top level of the experiment directories.
If someone wants to use NXRefine to analyze data collected as image
files, which are not stored in a directory tree compatible with the
above description, there are two options. Firstly, the NXBeamLine
class, which is described later, is designed to allow beamline-specific
methods of importing the data and metadata. These can be implemented in
separate packages that are imported into NXRefine as plugins.
Secondly, the image files can be loaded into HDF5 files using the
nexusformat command-line script,
‘nxstack’ and saved to the scan directories described above. Type
nxstack -h at the terminal command line to see possible options.