Server Management¶
NXRefine implements a data reduction workflow, which can be run as a series of line commands in the terminal. However, since some of the processes can take a long time to complete (from a few minutes to an hour, depending on the process and system being used), it is possible to queue these operations using the NXRefine’s queue manager, to be run locally using multiple cores or distributed to other nodes. The NXRefine queue manager can be configured to submit jobs to another job queue manager if one is available.
Server Directory¶
Here is the structure of the nxserver directory:
nxserver
├── nxserver.log
├── cpu1.log
├── cpu2.log
├── cpu3.log
├── settings.ini
├── nxsetup.sh
├── nxcommand.sh
└── task_list
├── info
└── q00000
└── locks
├── ...
└── ...
nxserver.log
This is a log file that records jobs submitted to the server queue.
cpu1.log, cpu2.log, …
These are log files that contain the output of jobs running on the server. The number depends on the number of simultaneous jobs that are allowed on the server, which is defined by the settings file.
settings.ini
A file containing default settings used by the NXRefine package, including server parameters, instrumental parameters, and parameters used in the data reduction workflow. When a new experiment is set up, a copy of these parameters is stored in the experiment directory (to be described later), so that they can be customized if necessary. These settings are described below.
nxsetup.sh
A shell script that could be used to initialize paths to the server directory or environment variables used by NeXpy. This could be run within a user’s
~/.bashrcfile, or by other shell scripts used to launch NXRefine workflow jobs (see below). Here is an example of what this file could contain.:export NX_LOCKDIRECTORY=/path/to/parent/nxserver/locks export NX_LOCK=10 export NX_SERVER=/path/to/parent/nxserverOther commands, e.g., to initialize a particular conda environment, could be also be added to this file.
nxcommand.sh
A shell script that is used if jobs need to be wrapped before submission to the job queue, e.g., using
qsub. Here is an example, in whichnxsetup.shis run in order to initialize NXRefine.:echo `date` "USER ${USER} JOB_ID ${JOB_ID}" source /path/to/parent/nxserver/nxsetup.sh <NXSERVER>
task_list
A directory that contains files that implement a file-based FIFO queuing system for server jobs.
locks
A directory that contains files that implement the nexusformat file-locking system. Locked files can be viewed, and removed if they are stale, using the “Show File Locks” dialog in the NeXpy “File” menu.
Note
The log files can be viewed using the “Manage Server” dialog and the settings file can be modified using the “Edit Settings” dialog, both of which are located in the “Server” menu in NeXpy.
Default Settings¶
The file, settings.ini in the server directory contains the default
settings for the server, the beamline, and the workflow. These values
can be changed, either by opening the “Edit Settings” dialog in the
NeXpy “Server” menu or at the command line using nxsettings -i,
which lists all the settings one by one, allowing their values to be
changed. Hitting the [Return] key keeps the current value.
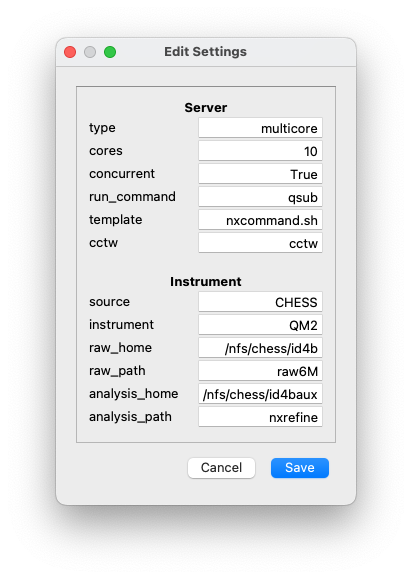
The figure shows an example of the first two sections of
settings.ini. The parameters in the first section are described
here. The other sections contain information concerning the location of
the data and default values of the data reduction parameters. They will
be described later.
Server Settings¶
The server settings are used by the workflow server, which is described in a later section. They define the server configuration, such as the number of simultaneous jobs that may be run, the command required to add them to the system’s standard job queues, and whether they need to be wrapped in a shell script.
- type
The server type can either be
multicoreormultinode. The only difference is that multinode servers have a list of defined nodes, to which jobs may be submitted, so their names will also be stored in the settings file. If jobs are submitted to a job server, without needing to specify the node, or if all the jobs are performed on the local machine, then the server type should bemulticore.- cores
This sets the number of jobs that can be run simultaneously by the server. Once reaching the limit, new jobs will only start as old ones are finished.
- concurrent
This determines whether parallelized processes should be used in the workflow. These speed up the computation, but can be disabled if they cause issues with the server. Note that this refers to whether multiple processes can be run simultaneously, e.g., in peaks searches, not whether multiple jobs can be submitted to the server. Valid values are
TrueorFalse.- run_command
This is a string that is prepended to any jobs that are submitted to the server. It can contain a set of switches in addition to the job submission command itself.
- template
In some systems, it is necessary to wrap the command that is submitted to the server in a shell script. This is the name of the script, which should be stored in the
nxserverdirectory. It should contain the string<NXSERVER>, which is replaced by the job command.- cctw
This is the path to the CCTW executable used to transform data from instrumental coordinates to reciprocal space.