Experiment Menu¶
The NXRefine plugin to NeXpy installs a top-level menu labelled “Experiment”. The sub-menus run operations to initialize the experiment layout, create experimental data templates, calibrate powder data, and initialize new data files.
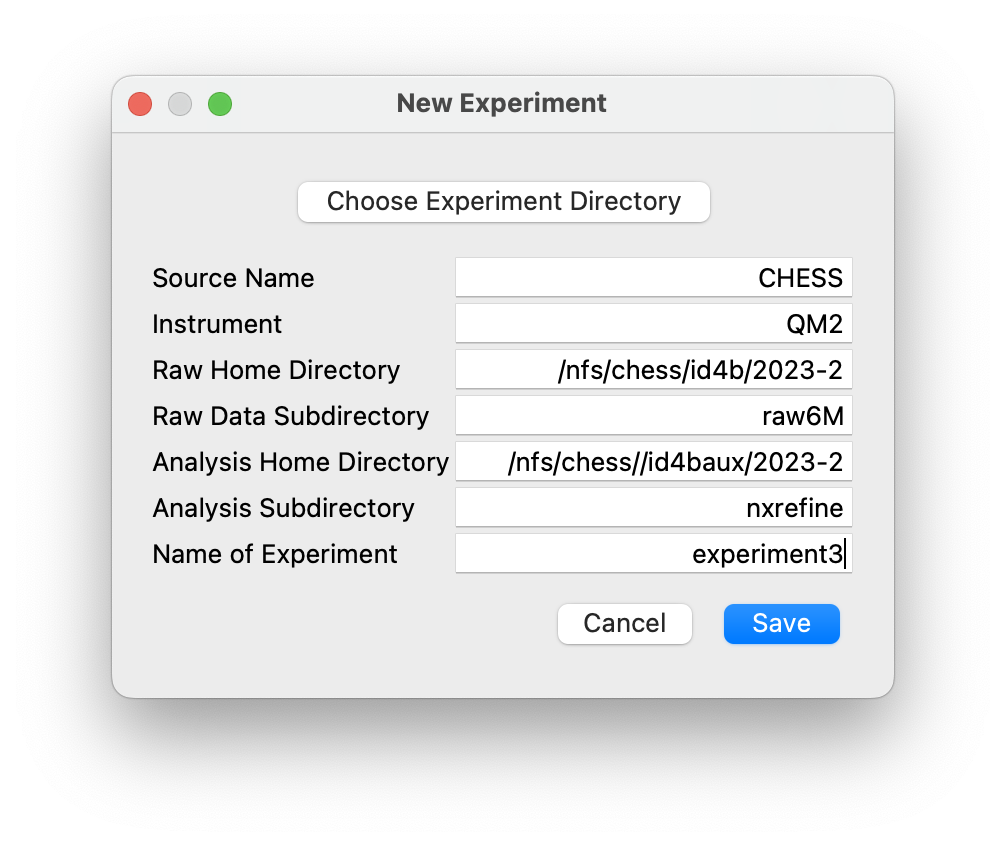
New Experiment¶
This dialog initializes a new experiment directory layout using the server settings to initialize default locations. When the dialog is launched, click on “Choose Experiment Directory” to launch the system file browser in order to select or create the new experiment directory.
There are two scenarios.
If
raw_homeis not blank in the server settings, the file browser will default to theraw_homedirectory, in which an experiment directory, containing the raw image files, should already exist. This experiment directory is then selected, after which the dialog above is created, with the experiment name (i.e., the base name of the experiment directory path) already filled in, along with the path to analysis home directory (analysis_homein the server settings) and the name of the analysis sub-directory if required. When the “Save” button is pressed, the new experiment directory is created within the analysis home directory if it does not already exist, and the experiment directory tree is initialized with thecalibrations,configurations,scriptsandtaskssub-directories.If
raw_homeis blank, the file browser will default to theanalysis_homedirectory, but another location can be selected if required. The file browser can be used either to select an existing experiment directory or to create a new one. The above dialog is then created with the experiment name given by the base name of the selected experiment directory path, and the analysis home directory defined by its parent. When the “Save” button is pressed, the experiment directory tree is initialized with thecalibrations,configurations,scriptsandtaskssub-directories.
A new settings.ini file is created in the tasks sub-directory,
with values copied from the equivalent file in the server directory,
excluding the “Server” section. This allows the refinement parameters to
be customized for each experiment.
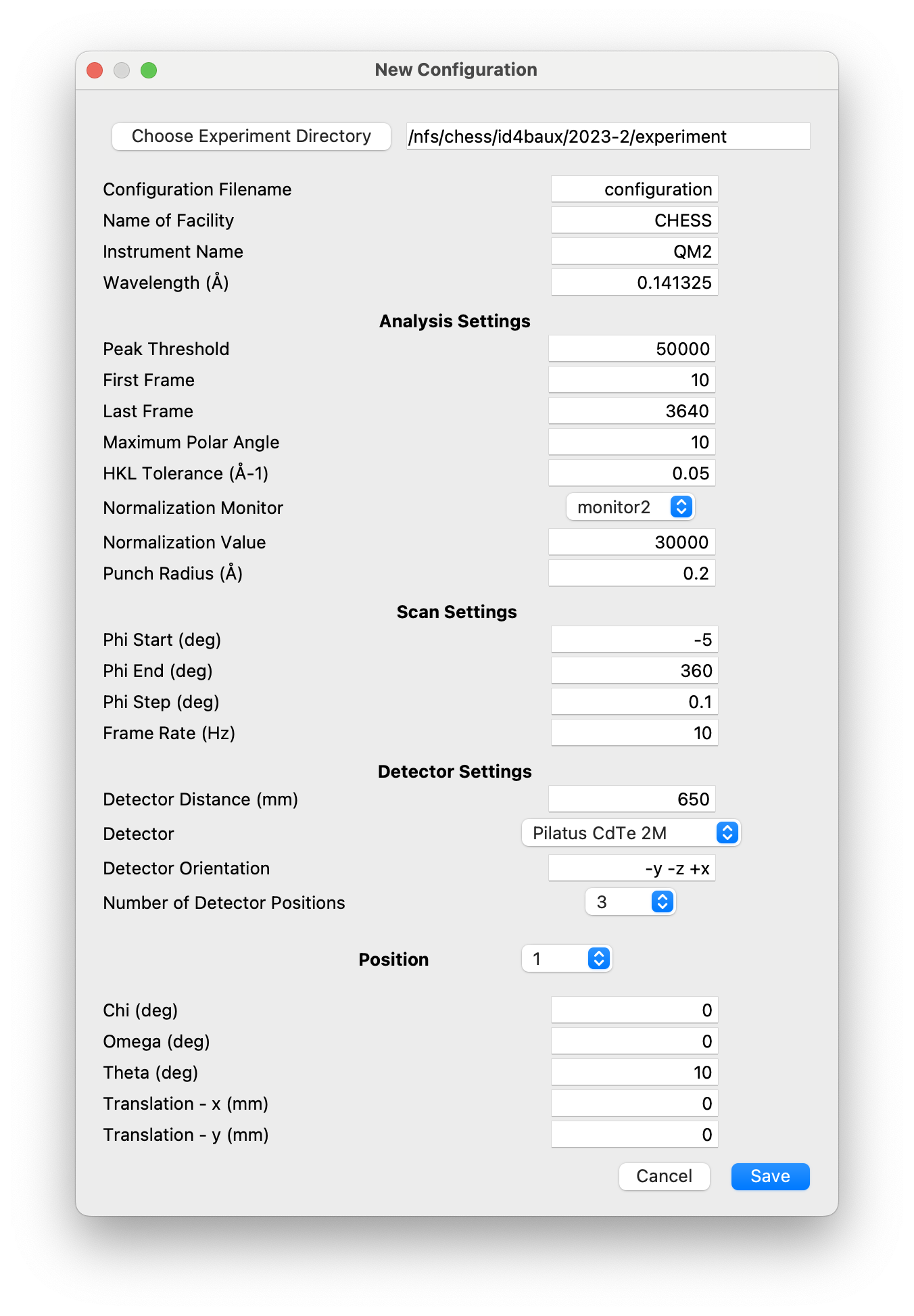
New configuration¶
This dialog creates NeXus files that are used as templates for the
experimental files that are used to store all the data and metadata
associated with a particular set of rotation scans. The initial metadata
is defined by parameters in the settings file in the tasks
sub-directory, which can be modified by the “Edit Settings” sub-menu
described below. However, some of the metadata will be refined using a
powder calibration, whose results are then stored in this file.
After selecting the experiment directory, the following dialog is created.
This allows the settings used in subsequent analysis to be initialized, the parameters defining the rotation scans (range, step size, frame rate) to be set, the detector configuration to be defined, and the angles and/or detector positions to be used in one or more rotation scans. These are all saved to the NeXus template. The wavelength and detector distance can be nominal values at this stage, since they are updated by performing a powder calibration. Similarly, the instrument angles, \(\theta\), \(\omega\), and \(\chi\) are set to the angles set by the motors, but will usually be refined when the sample orientation is determined.
It is possible to create more than one configuration template, if, for example, different angles and/or detector positions are used in different phases of an experiment. NXRefine allows the appropriate template to be selected when setting up the scan. A separate template should be created for each configuration that requires a change in the instrument calibration (wavelength, detector distance, detector translation) or scan angles.
The detector is chosen from a pull-down menu that contains all the detectors defined in the PyFAI package. This defines the number of pixels, their size, and a mask array used to exclude all the pixels within gaps between the detector chips.
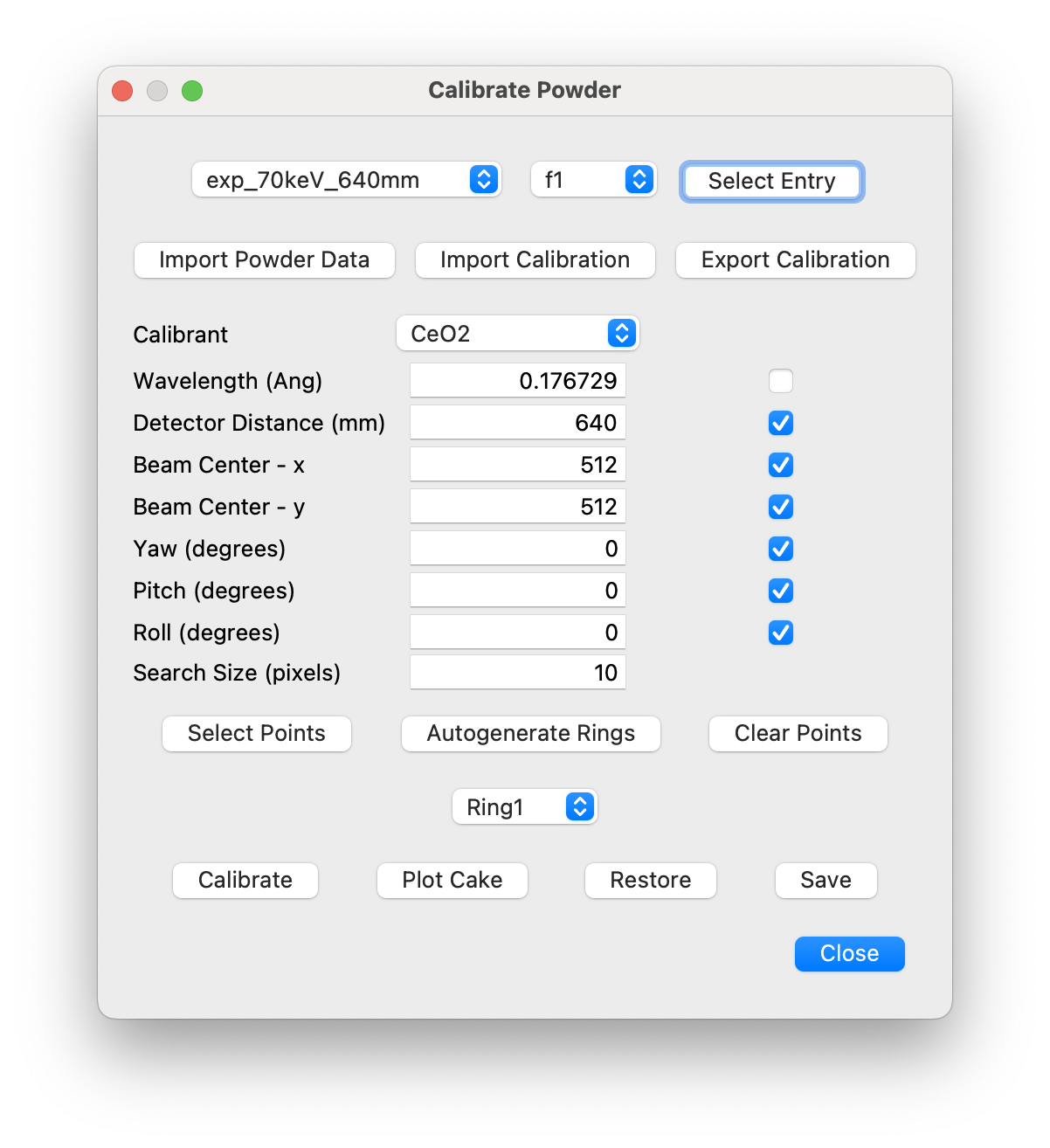
Calibrate Powder¶
This dialog will import a TIFF or CBF file containing measurements of a powder calibrant and refine the detector position and coordinates, using the PyFAI API. Alternatively, if the calibration parameters are already available in a PONI file, they can be directly imported. The resulting powder data and calbration parameters are then saved to the configuration template previously created using the New Configuration dialog.
After launching the dialog, select the entry in the configuration file to be calibrated by the powder measurement, i.e., the one with the correct wavelength, detector distance and translations. This expands the dialog with the default parameters defined by the settings file. The checkboxes at the side of each parameter specify whether the parameter is to be refined. By default, the wavelength checkbox is de-selected, since this is normally defined accurately by other means. It is too highly correlated to the detector distance for both to be refined simultaneously.
Then click on “Import Powder Data” to select the powder calibration file. This will generate a plot containing the powder data on a log scale. Select the approprate powder calibrant from those specified in the Calibrant pull-down menu.
If a PONI file already exists from a prior calibration, it can be imported using the “Import Calibration” button. If this is sufficiently accurate, it is not necessary to perform further calibrations. Instead the calibration parameters can be saved to the configuration file by clicking on “Save” and the dialog can be closed.
To obtain an initial calibration, zoom into this plot to display the first few rings.
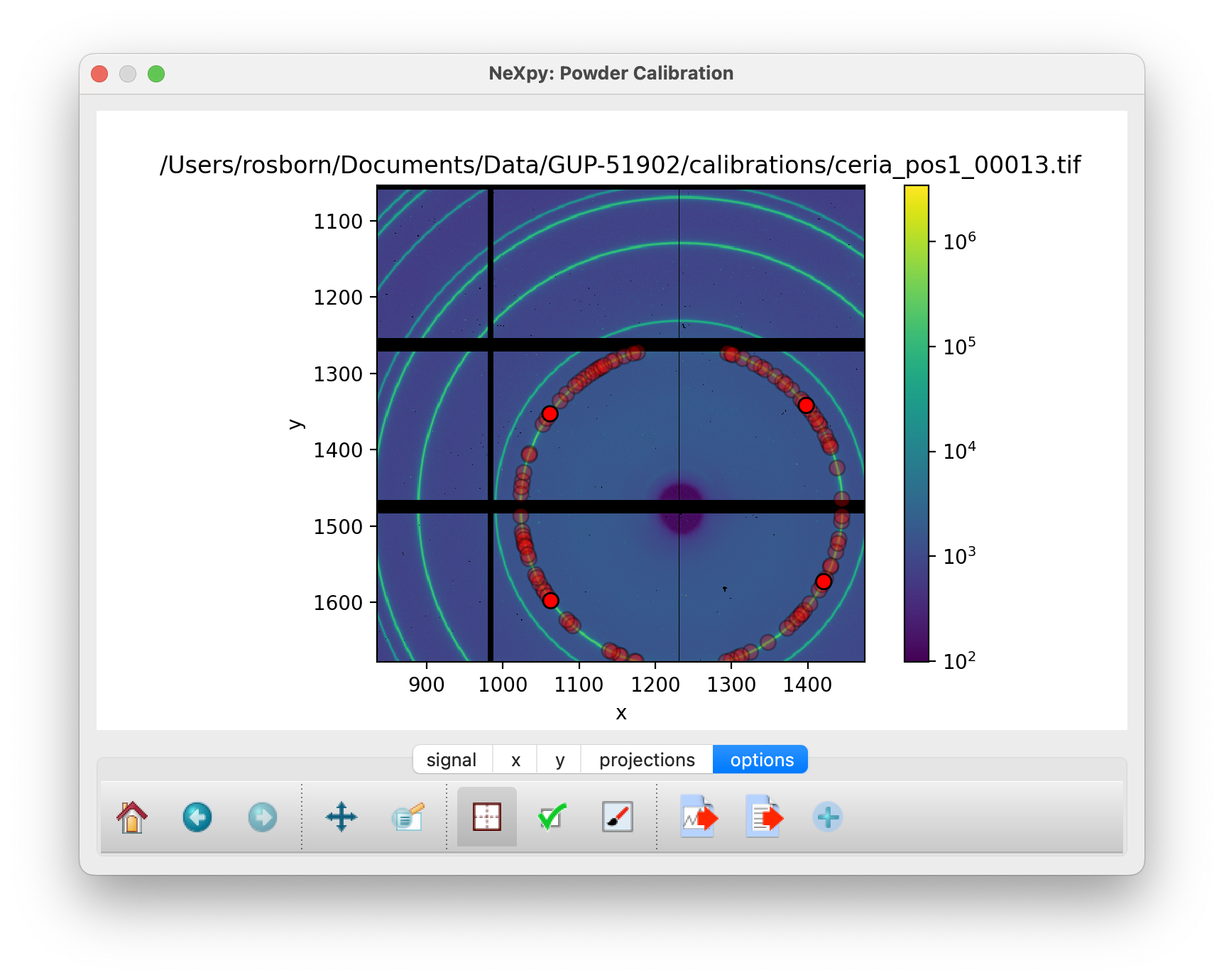
Points generated for the innermost ring after manually selecting four points¶
After clicking on “Select Points”, click somewhere on the innermost ring. This triggers the PyFAI Massif module, which automatically detects other points on the Debye-Scherrer ring that are contiguous to the selected point. Because of the gaps between detector chips, the Massif detection is confined to pixels within a single chip, so it is normally necessary to select other points on neighboring chips to complete a single ring. In the above ring, four selections, corresponding to the brighter red circles, were made.
It is only necessary to do this for a single ring. De-select the “Select Points” button and click “Calibrate” to perform an initial calibration. After this, it is possible to generate points automatically on the other rings using the “Autogenerate Rings” button. Select how many rings to generate, using the ring pull-down menu.
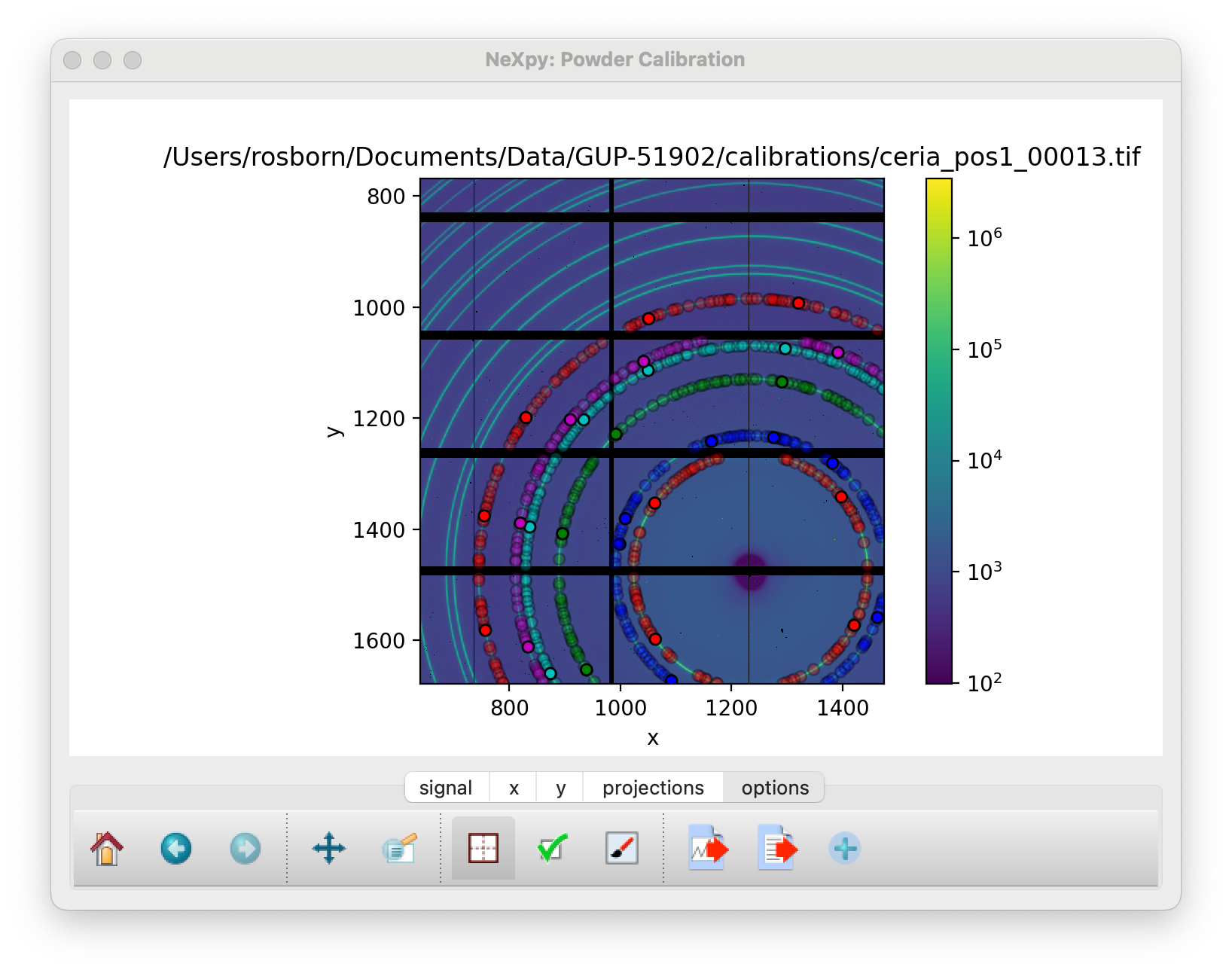
Autogenerated rings after selecting “Ring6” on the pull-down menu¶
When enough rings have been defined, click “Calibrate” again to produce a more accurate refinement.
The “Plot Cake” button can be used to generate a “cake” plot, in which all the powder rings, which are plotted against polar angle, should fall on vertical lines.
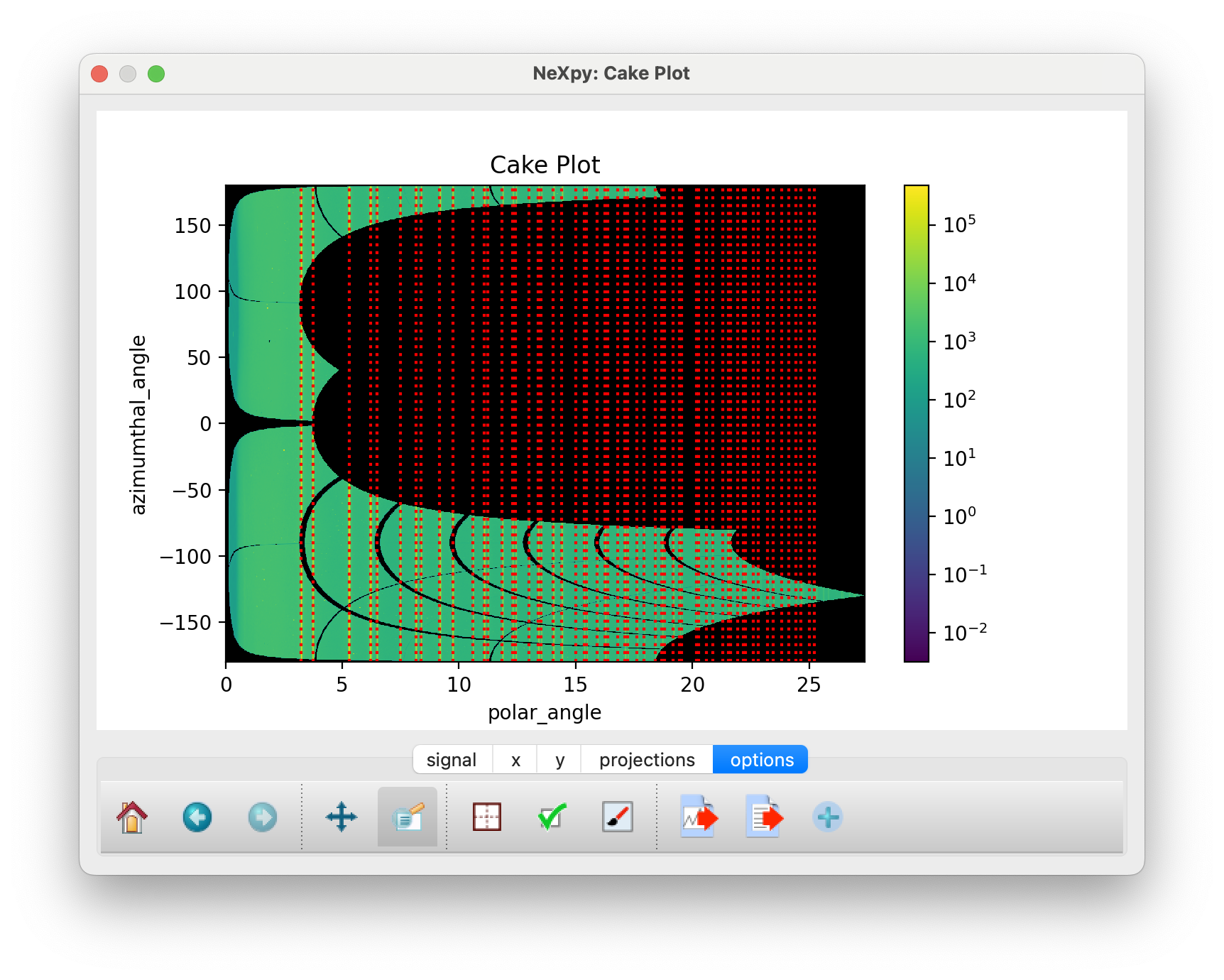
Cake Plot which allows a comparison of the powder data, plotted as a function of polar angle, with the theoretical powder lines (dotted red lines).¶
This can be used to determine whether the calibration is sufficiently good over the entire angular range of the detector. If there is evidence of distortions at higher polar angle, it may be necessary to autogenerate more rings before an additional calibration.
When the calibration is satisfactory, click “Save” to save both the powder calibration data and parameters to the configuration file. The calibration parameters can also be saved to a PONI file, using the “Export Calibration” button. This process should be repeated for each entry, after which the dialog can be closed.
Create Mask¶
This dialog creates a pixel mask that is used to exclude bad pixels from further analysis. As described above, when a new configuration file is created, a pixel mask that excludes gaps between detector chips is automatically added. Additional pixels can be excluded using this dialog, either by adding editable shapes that are constructively added to the existing mask or by importing the mask from an external file, which can store the mask in any image format. The latter is useful if a beamline regularly updates a particular detector’s mask as bad pixels are identified.
Warning
If an external mask is input using “Import Mask”, it will overwrite the existing mask. It is important therefore that the external pixel mask also excludes the detector gaps.
After launching the dialog, the current mask is automatically plotted, as an overlay on the powder diffraction data to enable the center of the beam and other features of the data to be identified.
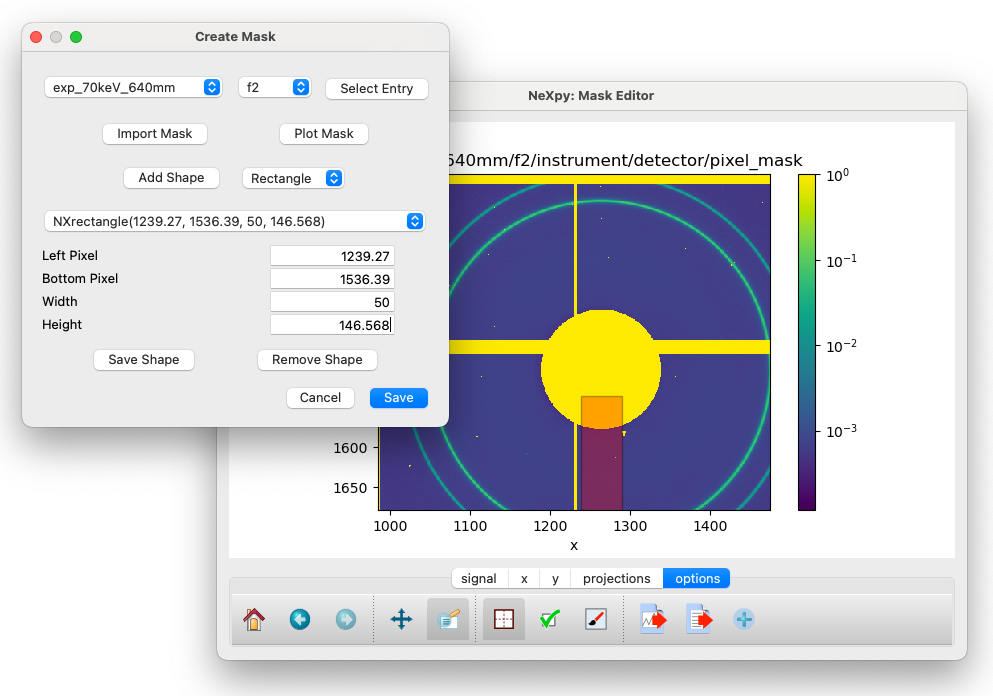
Create Mask dialog. The translucent shape shows the rectangle created by clicking “Add Shape”.¶
By clicking on “Add Shape” with either a rectangle or circle selected, a translucent shape is added to the plot. By default, it is centered on the beam center, but may be moved by dragging the center of the shape and/or resized by dragging one of the shape edges. When the shape has the correct position and size, click on “Save Shape” for the shape to be added to the current list. A pull-down menu allows existing shapes to be selected for further edits or removal
Note
After saving the shape, it is no longer draggable. However, the shape can still be modified by adjusting the shape parameters and then clicking on “Save Shape” again.
If a more complicated mask is required, it can be generated by an external image editor and imported using “Import Mask”.
When the mask is complete, click “Save” to save it to the configuration file.
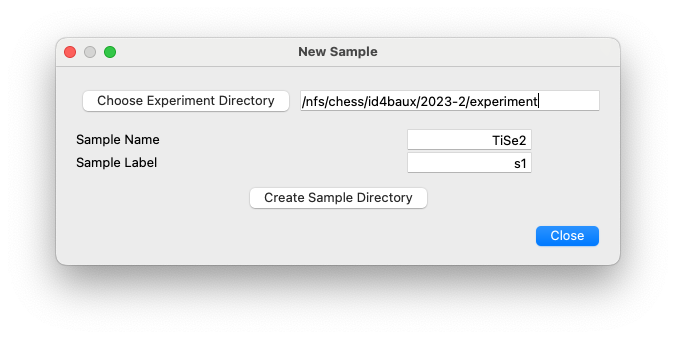
New Sample¶
This dialog has the single purpose of creating a directory tree for a new sample. The dialog enables the creation of a sample directory within the requested experiment directory and a sub-directory with a unique label for each instance of that sample measured during an experiment.
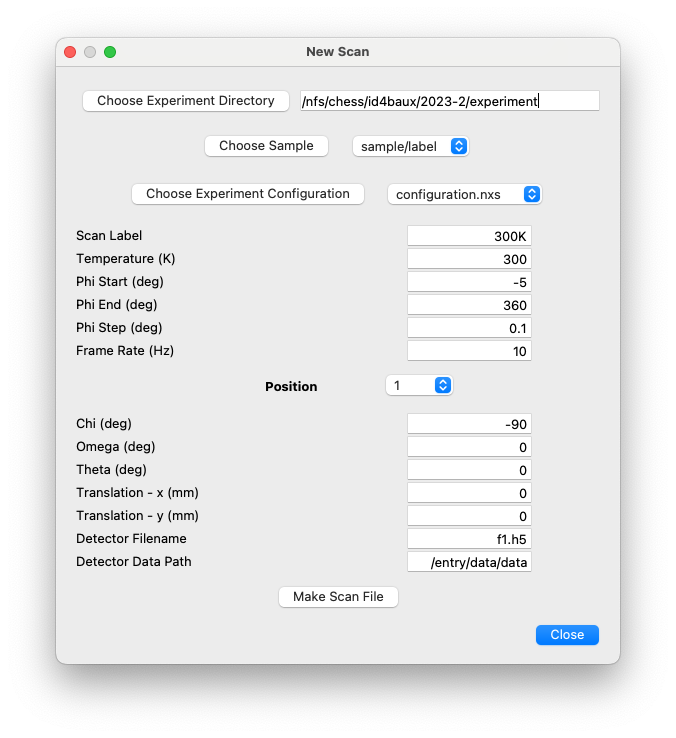
New Scan¶
This dialog is used to create a NeXus file in preparation for an experimental measurement. The file will be based on the selected configuration file and be saved in the specified sample/label directory. The name of the file will be “<sample>_<scan>.nxs”, where <scan> is the Scan Label specified in the dialog (‘300K’ in the image below).
The NeXus file is left open in the NeXpy tree. Multiple files can be created within the dialog, with different scan labels and, typically, different temperatures, before the dialog is closed.
External links to the raw data file are created in the NeXus file, even
if the data does not yet exist. In the example above, the external link
for the first detector position will be to f1.h5, in the <scan>
subdirectory. Similarly, the external link for the second detector
position would be to <scan>/f2.h5, etc. This experimental layout
is described in more detail in the `Experiment Layout`_ section above.
Import Scans¶
This dialog is for instruments in which the scans are already defined
using different methods to those above. For example, on the QM2
instrument at CHESS, the scans are defined in SPEC files, with the data
stored separately in a separate read-only directory. With this dialog,
the directories containing the raw images are associated with the
corresponding SPEC scan, allowing NeXus files to be automatically
generated. This customization is encoded in a QM2 sub-class of the
NXBeamLine class, which is installed separately as a NXRefine
plugin. The process for customizing other beamlines is described later.
Sum Scans¶
This dialog allows data in NeXus files collected under identical conditions to be summed to produce a single NeXus file that can be processed using the usual workflow.
Edit Settings¶
This dialog allows the settings, whose default values are defined in the
server directory (see Default Settings), to be customized for the
data reduction performed in the selected experiment. The settings are
stored in <experiment>/tasks/settings.ini. The meanings of each
setting are described in the next section.